Declustering & Compositing#
import geolime as geo
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from pyproj import CRS
geo.Project().set_crs(CRS("EPSG:26909"))
dh_tom = geo.read_file("../data/dh_tom.geo")
survey_tom = pd.read_csv("../data/survey_tom.csv")
Filtering on Tom West Prospect#
For simplicity of the tutorial, we will keep working only in the Tom West Prospect. The same workflow can be applied to all prospects.
dh_tom.set_region("TMZ", "Prospect == 'Tom West'")
dh_tom.keep_holes("TMZ", method=geo.RegionAggregationMethod.ALL)
dh_tom.plot_2d(property="Prospect", agg_method="first", interactive_map=True)
Declustering#
sns.histplot(
x=dh_tom.data(
"Zn_pct",
region="Zn_pct.notna()"
),
bins=30,
log_scale=True,
stat='percent',
color='red'
)
<Axes: ylabel='Percent'>
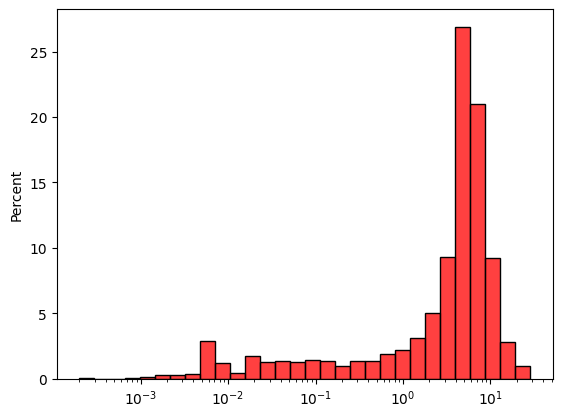
Using fairly the size of the drill spacing along x and y and using a large scale in z, we can compute the declustering weights.
declus_weight_mw = geo.moving_window_declus(
obj=dh_tom,
obj_region="Zn_pct.notna()",
obj_attribute="Zn_pct",
diam_x=200,
diam_y=200,
diam_z=1500,
geometry="BALL"
)
print("Weight mean: ", declus_weight_mw.mean())
Weight mean: 1.0
moving_window_declus outputs a numpy array of values with a mean of 1. The declustered histogram can also be visualised using seaborn and the weights parameter.
declus_weight_mw
array([0.77649391, 0.79988228, 0.8014917 , ..., 0.54269942, 0.54269942,
0.5412247 ])
dh_tom["declus_weight_cd"] = "nan"
dh_tom.update_property("declus_weight_cd", data=declus_weight_mw, region="Zn_pct.notna()")
fig, ax = plt.subplots()
sns.histplot(
x=dh_tom.data("Zn_pct", region="Zn_pct.notna()"),
bins=30,
log_scale=True,
stat='percent',
ax=ax,
color='red',
label='Clustered'
)
sns.histplot(
x=dh_tom.data("Zn_pct", region="Zn_pct.notna()"),
weights=declus_weight_mw,
bins=30,
log_scale=True,
stat='percent',
ax=ax,
color='green',
label='Declustered'
)
ax.legend()
plt.show()
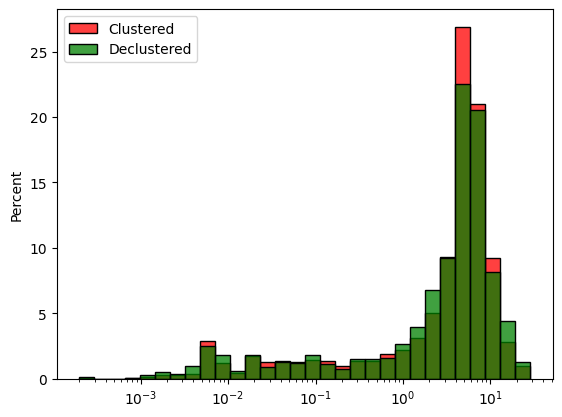
Cell Declustering is also available and works in the same manner.
declus_weight_cd = geo.cell_declustering(
obj=dh_tom,
obj_region="Zn_pct.notna()",
obj_attribute="Zn_pct",
size_x=200,
size_y=200,
size_z=1500,
nb_off=50
)
print("Weights mean: ", declus_weight_cd.mean())
Weights mean: 1.0
fig, ax = plt.subplots()
sns.histplot(
x=dh_tom.data("Zn_pct", region="Zn_pct.notna()"),
bins=30,
log_scale=True,
stat='percent',
ax=ax,
color='red',
label='Clustered'
)
sns.histplot(
x=dh_tom.data("Zn_pct", region="Zn_pct.notna()"),
weights=declus_weight_cd,
bins=30,
log_scale=True,
stat='percent',
ax=ax,
color='green',
label='Declustered'
)
ax.legend()
plt.show()
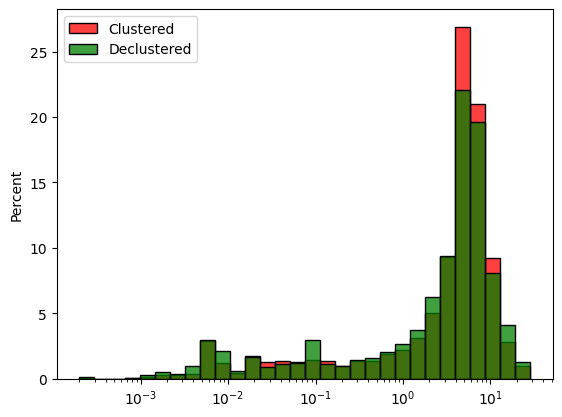
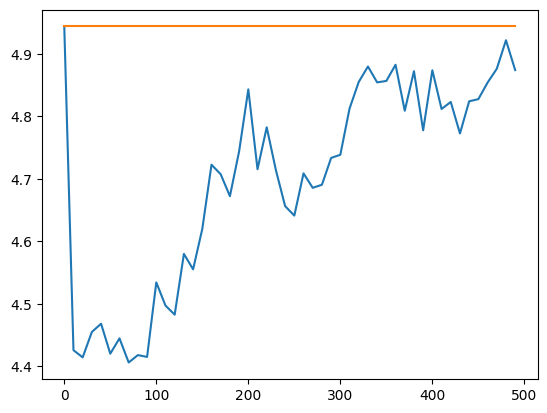
Declustering size of cell or ellipsoid can be found using a loop over different size and taking the size minimizing the mean of the data.
x_cs_cd = np.arange(0, 500, 10)
naive_mean = dh_tom.data("Zn_pct", "Zn_pct.notna()").mean()
var_mean_cd = [naive_mean]
for s in x_cs_cd[1:]:
declus_weight_cd = geo.cell_declustering(
obj=dh_tom,
obj_region="Zn_pct.notna()",
obj_attribute="Zn_pct",
size_x=s,
size_y=s,
size_z=1500,
nb_off=25
)
var_mean_cd.append(np.mean(declus_weight_cd * dh_tom.data("Zn_pct", "Zn_pct.notna()")))
fig, ax = plt.subplots()
ax.plot(
x_cs_cd,
var_mean_cd,
label="Declustered Mean"
)
ax.plot(
x_cs_cd,
np.full((len(x_cs_cd)), naive_mean),
label="Naive Mean"
)
plt.show()
Compositing#
To perform compositing, we can create a new property called sample_length and analyse its distribution to find the most common lenght of the samples.
dh_tom.set_property(name="sample_length", data="TO - FROM")
dh_tom.user_properties()
['X_COLLAR',
'Y_COLLAR',
'Z_COLLAR',
'X_M',
'Y_M',
'Z_M',
'X_B',
'Y_B',
'Z_B',
'X_E',
'Y_E',
'Z_E',
'Ag_ppm',
'Pb_pct',
'Zn_pct',
'BD_tonnes_m3',
'Method',
'Comment',
'LowRecovery___85pct',
'HoleType',
'Year',
'Property',
'Prospect',
'Grid',
'NewsRelease',
'Length_m',
'declus_weight_cd',
'sample_length']
dh_tom.describe(properties=['sample_length'])
| sample_length | |
|---|---|
| count | 2668.000000 |
| mean | 1.070956 |
| std | 0.417748 |
| min | 0.100000 |
| 25% | 0.920000 |
| 50% | 1.000000 |
| 75% | 1.500000 |
| max | 4.600000 |
geo.histogram_plot(
data=[{"object": dh_tom, "property": "sample_length"}],
width=650,
height=400,
histnorm='percent',
title=f'Sample length {len(dh_tom["sample_length"])}'
)
1m seems to be the most common value and will be used for compositing. Additional check such a histogram comparison can be made to asses the composite length.
dh_tom_2 = geo.compositing(
drillholes=dh_tom,
attribute_list=["Zn_pct", "Pb_pct", "Ag_ppm", "declus_weight_cd"],
composit_len=1.,
minimum_composit_len=0.3,
residual_len=0.5,
survey_df=survey_tom
)
2026-02-05 11:17:26,963 [INFO] GeoLime Project - |GEOLIME|.drillholes_compositing.py : Following survey mapping has been identified: {
"AZIMUTH": "Azimuth",
"DEPTH_SURVEY": "Depth_m",
"DIP": "Dip",
"HOLEID": "HoleID"
}
fig, ax = plt.subplots()
sns.histplot(
x=dh_tom.data("Zn_pct", region="Zn_pct.notna()"),
bins=30,
log_scale=True,
stat='percent',
ax=ax,
color='red'
)
sns.histplot(x=dh_tom_2.data("Zn_pct", region="Zn_pct.notna()"), bins=30, log_scale=True, stat='percent', ax=ax, color='green')
<Axes: ylabel='Percent'>
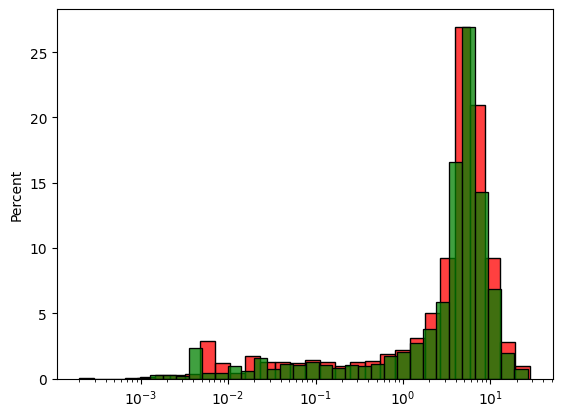
DataFrame can also be created to have a summary comparing the original and composited Drillholes.
pd.DataFrame(
[dh_tom.describe("Zn_pct"), dh_tom_2.describe("Zn_pct")],
index=['original', 'composited']
).T
| original | composited | |
|---|---|---|
| count | 2665.000000 | 2965.000000 |
| mean | 4.943900 | 4.901401 |
| std | 4.011429 | 3.752160 |
| min | 0.000200 | 0.000900 |
| 25% | 1.800000 | 2.178125 |
| 50% | 4.800000 | 4.824430 |
| 75% | 6.700000 | 6.600000 |
| max | 28.600000 | 26.537500 |
Export to file for future use. geo files allow faster reading and writing.
dh_tom_2.to_file("../data/dh_tom_comp.geo")